Fall semester 2025
Field-based environmental DNA analysis protocols with CRISPR-Dx enable to rapidly detect elusive species
Flurin Leugger
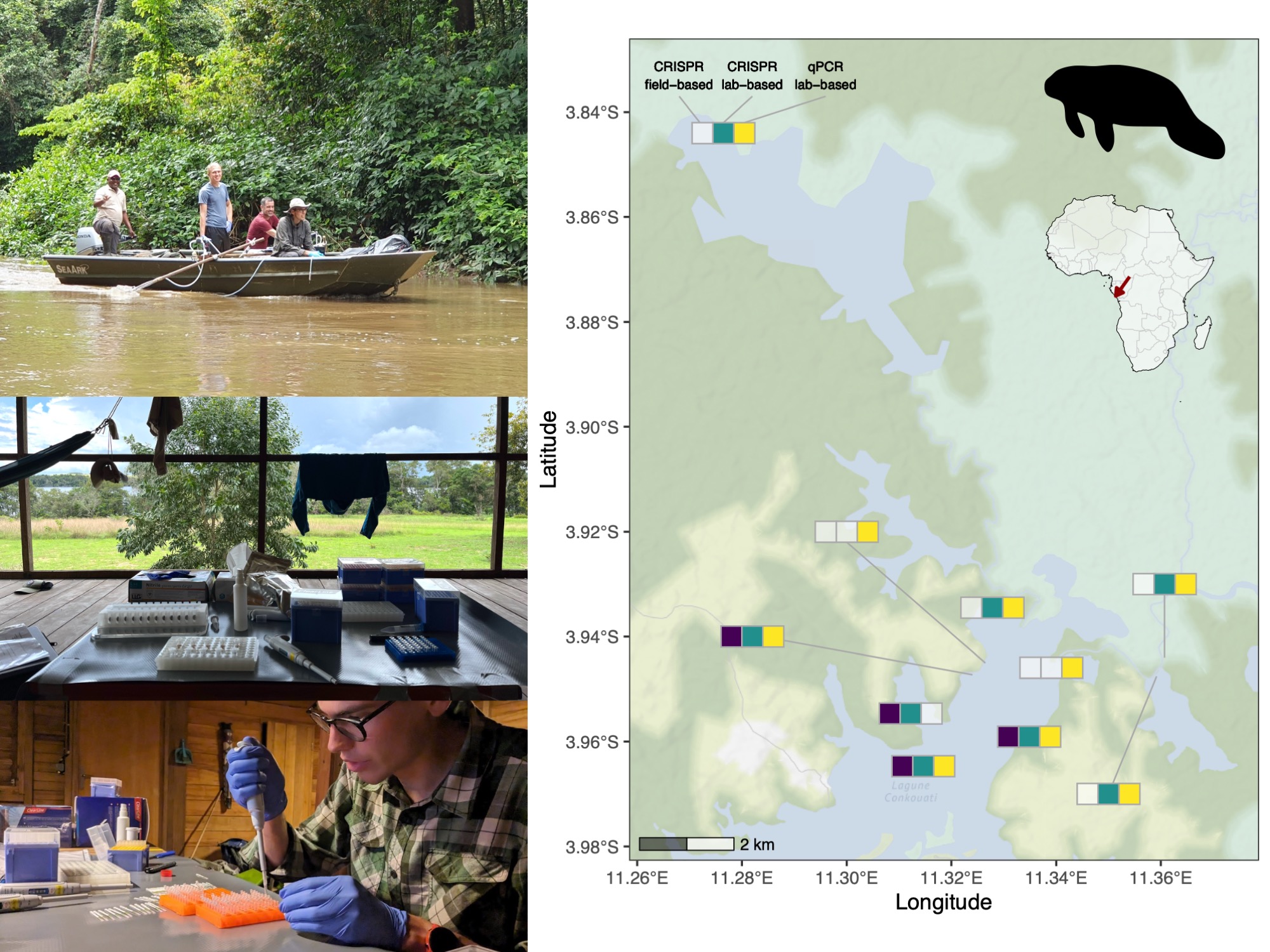
Environmental DNA workflow: taking water samples in the Conkouati-Douli National Park (Republic of the Congo; top left; © Nina Giotto), extracting the DNA at the camp in the national park (middle left; © Flurin Leugger) and analysing the samples using CRISPR-Dx assays with lateral flow strips (bottom left; © Michel Schmidlin). We successfully detected African manatee DNA with the field-based method, though the lab-based method were more sensitive, resulting in additional detections.
Environmental DNA (eDNA) has emerged as a promising method for biodiversity assessment, but current protocols are often slow and reliant on advanced laboratory infrastructure. The application of eDNA is currently mainly limited to well-developed countries and, for example, rarely applied in tropical regions. Therefore, we have developed a field-deployable eDNA protocol using CRISPR-based diagnostics and lateral flow tests, enabling rapid, on-site detection of species. To demonstrate its application, we created a CRISPR-Dx assay targeting the African manatee (Trichechus senegalensis), arguably one of Africa’s least studied mammals. We successfully detected its DNA in water samples from a national park in the Republic of Congo analyzing the samples in a hut in the national park. These field-based detections were later validated in the lab using a lab-based CRISPR-Dx protocol and quantitative PCR (qPCR) assay. While less sensitive than lab-based methods, the field protocol offers significant advantages in speed, cost, and accessibility. This field-based eDNA analysis protocol with CRISPR-Dx and lateral flow tests as read out has the potential to expand the reach of eDNA monitoring and support conservation efforts for elusive and threatened species.
___________________________________________________________________